
Wageningen World
CRISPR-Cas pioneer John van der Oost: ‘Chance has been a determining factor in my scientific career’
When John van der Oost arrived in Wageningen all those years ago, no one had heard of CRISPR-Cas – not even him. His path to discovering this revolutionary technique was determined in part by random chance and in part by seizing the opportunities that arose. Van der Oost will be retiring this year.
Text: Albert Sikkema | Photography: Eric Scholten
The path that led microbiologist John van der Oost to the discovery of the genetic cut-and-paste technique of CRISPR-Cas was long and winding. ‘Chance has been a determining factor in my scientific career,’ he says. That started with his choice of degree subject. ‘Just before my final school exams, I read a chapter in our biology textbook that the teacher had skipped. It was about molecular biology and how organisms store information in their DNA and transcribe it into proteins via RNA.'
Wageningen World
'I found that so fascinating that I decided to do a degree in biology. At that time, I was a member of the PSP, the Pacifist Socialist Party, so the obvious choice was to go to the more progressive University of Amsterdam. Funnily enough, I thought that the VU, the Free University of Amsterdam, was the progressive one, so I enrolled there. I discovered that some of the lecturers started their lectures with passages from the Bible. Even so, it felt like a really good place, so I stayed.’
Unlike most of his friends, he chose not to specialize in research on animals. ‘I had become a vegetarian and I didn’t want to kill animals, not even for research. I wanted to do courses in plant physiology, but they were already full. I was told to try microbiology.’
Van der Oost found it interesting and he started investigating energy production by bacteria. In 1984, he was taken on as a PhD candidate at VU University Amsterdam, studying how cyanobacteria convert sunlight into hydrogen. ‘That was pioneering work. It turned out that the lab in Amsterdam didn’t have the right cyanobacteria so I hitchhiked to Paris with my girlfriend Paulien. I got the cyanobacteria I needed from the Pasteur Institute’s collection of cultures, and carried them back to Amsterdam in test tubes I put in my rucksack.’
Nowadays, you can analyse a gene in a couple of days, but back then it took me a year
Van der Oost takes his PhD thesis from the shelf. In it, he not only describes the conditions under which the bacteria start making hydrogen but also identifies a key gene responsible for this. He opens the thesis at the relevant pages, full of DNA code – the letters G, A, T and C. ‘Look, this is the gene. Of 1500 base pairs, no less. Nowadays, you can analyse a gene in a couple of days, but back then it took me a year. When I started, my supervisors told me I’d never manage it.’
After his application for a grant to do follow-up research in Brighton was rejected, he ended up via a roundabout route – six months at the University of Helsinki, two and a half years at an institute in Heidelberg – back at the VU University in Amsterdam. But not for long. The head of the Microbiology group, who was planning to get him a permanent contract, died suddenly from a heart attack in the lab. His successor had other plans, so that was the end of that job. ‘It was quite a blow because by then I was married to Paulien and we had three children.’ He applied for three microbiologist jobs at different places in the Netherlands, and was taken on by Professor Willem de Vos in Wageningen.

When Van der Oost arrived in Wageningen 30 years ago, aged 37, no one had heard of CRISPR-Cas, including him. He was appointed head of the Bacterial Genetics research group. ‘For the first ten years, we focused on how archaea and bacteria, relatively simple microorganisms without nuclei, evolved to become eukaryotes, the organisms ranging from fungi to humans that do have nuclei. In particular, we investigated which proteins in archaea ended up in eukaryotes too. That gave some nice research results and I got a prestigious Vici grant from the Dutch Research Council in 2005. That let me expand this line of research.’ Shortly afterwards, he was promoted to a position as professor holding a personal chair. ‘Thijs Ettema was one of the students on my microbial evolution course. Thijs later started his own group in Sweden, where he found some important pieces of the puzzle of the evolution of the prokaryotic archaea into the more complex eukaryotic life. He recently returned to Wageningen and now he is officially my boss – isn’t that great?’
When did CRISPR-Cas come into the picture?
‘Thijs Ettema obtained his doctorate in September 2005. One of the members of the thesis committee was the bio-informatics specialist Eugene Koonin. He told us for the first time about Clusters of Regularly Interspaced Short Palindromic Repeats (CRISPR), repeated pieces of DNA in bacteria genomes. We’d found these CRISPR pieces in 2001 in a microbial genome sequence, but had no clue about their function. The answer was provided by the Spanish scientist Francisco Mojica, who discovered that the variable DNA pieces in CRISPR were often identical to pieces of DNA in bacteriophages, viruses that use bacteria as their hosts. He proposed that CRISPR could be part of a bacterial immune system to protect against bacteriophages. Koonin also said his group had discovered clusters of genes linked to these CRISPR sequences, that together were probably responsible for a novel immune system such as Cas proteins.
Together with a postdoc in my group, Stan Brouns, who became a professor at Delft last year, I decided to try and figure out the mechanism of CRISPR-Cas. So I changed my Vici project completely to focus on CRISPR-Cas rather than archaea. Later, I sent the Dutch Research Council, which had funded the research, a letter admitting I had used most of the grant to study something completely different. I never got a reply, so I assume they agreed it was a good decision.’
What was the result?
‘In 2008, we described the first mechanistic insights of CRISPR-Cas in an article in Science. That was a real breakthrough and certainly one of the most spectacular publications in my career. The article is considered a milestone in CRISPR research and has been cited over 3000 times.’
When did it change from fundamental research on bacteria into a tool for editing DNA?
‘Back then, the dairy company Danisco had a research team that was trying to understand how lactic acid bacteria protect themselves against bacteriophages. They did an experiment in which they added a bacteriophage to a reactor with lactic acid bacteria. The bacterial culture collapsed because the bacterial cells were killed by the viruses. But after one day the bacterial culture started to grow again. That was because a few bacteria had developed resistance to the bacteriophage.
When the researchers analysed the CRISPR DNA of the surviving lactic acid bacteria, they found pieces of the viral DNA had been incorporated. That adaptation allowed the bacterium to survive in the presence of the virus. When the researchers removed that piece of viral DNA from the CRISPR, the bacterium became susceptible to the virus again. This was proof that the CRISPR system is an adaptive immune system that can recognize and neutralize viruses in a very targeted way. The big question was: how did it work?
After Koonin’s visit, we had decided to study the CRISPR-Cas system of the model bacterium E. coli. Unfortunately, we didn’t find a match with the DNA of known bacteriophages in the CRISPR DNA of the model bacterium. So we decided to insert pieces of a specific E. coli bacteriophage into the CRISPR DNA ourselves. That meant we’d created the first designer CRISPR, which we went on to test.
When we added the bacteriophage to a culture medium of unmodified E. coli bacteria, the lawn of bacteria contained clear zones that corresponded to areas where bacteria did not survive the viral infection. When we added the bacteriophage to our modified designer E. coli, we didn’t see any holes. That was the eureka moment: we’d been able to give a bacterium highly specific protection against a bacteriophage. We called it a “flu shot for bacteria”. In the years that followed, various research groups worked out the complete mechanism behind the CRISPR-Cas system, step by step.’
When did it become a business?
‘Stan Brouns thought our CRISPR system could be used more broadly, as a new instrument for genome editing in which the DNA of plants and humans is edited deliberately and in a targeted way. Together, we explained the potential of CRISPR-Cas to WUR’s patent expert, Paul van Helvert. Then the three of us went to a patent bureau in London to discuss the options. In 2011, we submitted a patent application describing how we could make targeted changes in any DNA, including the DNA of plants or humans.’
You were involved from the start in research that was later rewarded with the Nobel Prize.
‘Emmanuelle Charpentier and Jennifer Doudna, who got the Nobel Prize for Chemistry in 2020 for the development of the CRISPR-Cas9 technique, described the same mechanism we did in 2012, one year after we had filed our patent. However, they used a different system, the Cas9 protein of a lactic acid bacterium. That system is very compact and straightforward whereas our Cascade system consists of over a dozen proteins, making it much more complex and harder to use as a general tool for genome editing.’
So why did you apply for patents?
‘Of course we didn’t know in 2011 that Cas9 would work so much better. The organization was also encouraging us to protect our findings and ideas. A patent application is always speculative; whether it will be a good investment is pretty unpredictable. But a patent is also regarded as a publication that is good for your CV and could strengthen research proposals. So it was a strategic decision to file patents to protect these exciting ideas. Over the past 15 years, I’ve submitted more than 30 patents.’
What have you made from them?
‘The revenue from most patents is less than the costs. Still, a few were successful, especially our patent on Cas12. How that came about is a nice story. At a conference in Paris, I spoke to one of the aforementioned Koonin’s employees. They said, “We’ve discovered a new CRISPR system, Cas12. You should do some work on it.” After a few months, we had our first results. It turned out Cas12 is quite different from Cas9. I decided to submit a project proposal so I could recruit more people to work on this. But to do that, I would have to write down our unpublished work and our plans. So the patent expert Van Helvert and I decided to submit a patent application, even though we still needed to figure out an awful lot of details about Cas12.
A few months later, I met Feng Zhang, a molecular biologist from the Broad Institute in Boston, at the annual CRISPR meeting. He turned out to have got the same tip from Koonin and was therefore also working on Cas12. We decided then to collaborate on this. Zhang’s team had already advanced a long way and the Broad Institute had applied for a patent too, only six weeks after us. We soon agreed to work together and manage our patents jointly.’
What is Cas12 used for?
‘Cas12 can be used to cure certain diseases in humans. A great example is curing one form of chronic anaemia, caused by a genetic mutation in the haemoglobin in the red blood cells. Cas12 can repair the blood cells by editing a piece of DNA. Furthermore, Cas12 is being used to make plants resistant to diseases.’
Who gets the patent revenue?
‘Some of the money goes to the inventors, some to the Microbiology chair group and the rest to WUR’s Agrotechnology & Food Sciences group (AFSG). Willem de Vos and I agreed that the latter part would be put in a fund called the Innovation Platform for Microbiology. Every now and then, AFSG researchers can apply for money from the fund to set up microbiology-related innovative research. It’s a revolving fund, which is wonderful.’
Do you have other patents that are now being used?
‘In 2015, we did research on bacteria that can break down the cellulose in plant waste, which would make the waste easier to recycle. We found two candidates in a compost heap not far from Wageningen. When we analysed the genomes of these bacteria, we found an alternative Cas9 system. We applied for a number of patents for this too.
A few months later, I saw Christa Testerink, the professor of Plant Physiology at Wageningen, on a TV science programme talking about her work at Wageningen and at the International Rice Research Institute (IRRI) in the Philippines on salt-tolerant rice crops. Then I thought: why don’t we make these patents available free of charge to non-profit projects like this so they can modify plants to cope with climate change? That needed the consent of the university’s Executive Board as they are the official patent holder. Fortunately the then Board President Louise Fresco was enthusiastic as well. We launched the plan at the start of the academic year in 2021.’
Do you already have results?
‘Christa and I have received money from donors through University Fund Wageningen to start a project with the IRRI. Christa’s group recently managed to use our optimized system to modify the DNA of rice plants for the first time. We have to edit several genes to increase the salt tolerance of rice plants. We’ll shortly be sharing this knowledge with the IRRI.’
When the patent was released in 2021, Extinction Rebellion (XR) organized protests in the Aula and Hotel De Wereld. You wanted to talk with them, but they wouldn’t say anything. You got very angry.
‘Yes. As researchers, we’re often accused of using our knowledge to help companies. Now we were giving our knowledge away for free to make the world a better place and they were still demonstrating. I found that so frustrating, especially since I often agree with XR, and I sometimes feel like glueing myself to the A12 motorway in aid of a better climate. So I genuinely wanted to talk to them about how our technology and our knowledge could improve the world. But I couldn’t. That was very disappointing.’
Technology and patents are not automatically bad
‘Technology and patents are not automatically bad; it all depends on what you do with them. You can use the technology to sustainably increase food production. I don’t think I’ve changed much in that respect since I went round Amsterdam sticking up PSP posters.’
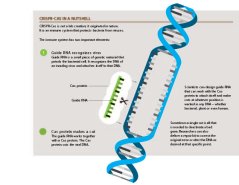
CRISPR-Cas in a nutshell
CRISPR-Cas is not a lab creation; it originated in nature. It is an immune system that protects bacteria from viruses.
The immune system has two important elements:
Guide RNA recognizes virus
Guide RNA is a small piece of genetic material that patrols the bacterial cell. It recognizes the DNA of an invading virus and attaches itself to that DNA.
Cas protein makes a cut
The guide RNA works together with a Cas protein. The Cas protein cuts the viral DNA. Sometimes a single cut is all that is needed to deactivate a bad gene. Researchers can also deliver a repair kit to correct the original error or alter the DNA as desired at that specific point. Scientists can design guide RNA that can work with the Cas protein to attach itself and make cuts at whatever position is wanted in any DNA – whether bacterial, plant or even human.
John van der Oost (67)
In addition to outreach activities, he is a scientific adviser to the companies NTrans Technologies, Scope Biosciences and Hudson River Biotechnology.
1984: MSc in Molecular Biology, VU University Amsterdam
1989: Promotie VU
1995: Laboratory of Microbiology, WUR
2005: Professor holding a personal chair in Bacterial Genetics, WUR
2008: First CRISPR-Cas publication in Science
2013: Member of the European Molecular Biology Organization
2017: Member of the Royal Netherlands Academy of Arts and Sciences
2018: Winner of the Spinoza Prize
2025: Winner of the M.W. Beijerinck Virology Prize
